微单倍型是一种新型法医遗传学分子标记,可用于个体识别、祖先推断和混合样本DNA的检验。虽然现已有微单倍型基因座的标准命名方法,但简明的微单倍型等位基因命名规则尚未见到。微单倍型遗传标记相对于单核苷酸多态性(single nucleotide polymorphism, SNP)的优势在于其多等位基因。一般来说,微单倍型基因座包含的SNP数目越多,其有效等位基因数(effective number of alleles, Ae)和信息含量(informativeness, In)就倾向于越高。这些基因座更适宜于法庭科学应用,因其复杂的等位基因更能满足应用需求。因此,为更便于应用,我们建议使用阿拉伯数字命名微单倍型等位基因。具体规则如下:以人类基因组正链作为比对微单倍型,GRCh38作为参考序列。将组成微单倍型的SNP根据它们在人类基因组中的物理位置进行排序,dbSNP数据库中的参考等位基因则作为这些SNP可能出现的备选基因型。先按照参考等位基因列出所有可能出现的微单倍型等位基因,然后按照字母表顺序排序,并从1开始以连续正整数依次命名排序后的等位基因。稀有的微单倍型等位基因仍然用SNP分型的组合命名,列在基因座内所有用数字命名的等位基因之后。本文使用9947A、2800M和两份志愿者DNA制备了1∶2∶4∶8比例的DNA混合物,对单一来源样本和混合物进行微单倍型测序、数据分析,并对等位基因进行基因型组合命名和数字化命名两种方式的对比展示。本文建议的数字化命名法其优势在于:首先,这种命名法使得微单倍型的法庭科学应用更便捷,可避免使用SNP分型组合的复杂命名方式,在混合DNA分析中优势尤其显著。其次,在每个微单倍型基因座内部,可以按照阿拉伯数字的顺序排列等位基因,从而能为微单倍型数据的展示和交流提供统一的等位基因排序方式。第三,数字化的等位基因命名方式更易为法医DNA技术人员接受,因为这与法庭科学STR等位基因的命名方式类似。第四,当前的群体遗传学软件,如PowerStats、Arlequin、STRUCTURE等,只接受数字作为导入的等位基因名称,该命名方法能更好地与这些既有软件衔接关联。因此,数字化的微单倍型等位基因命名法应能为法医学应用与实践带来便利。
First author: SONG Jiaojiao, female, Jinzhong of Shanxi, master of science, mainly focusing on the detection of biochemistry and molecular biology. E-mail: 1248390463@qq.comk
Microhaplotypes, an emerging type of forensic genetic marker, have been being used for individual identification, ancestry inference and mixture deconvolution as they were incessantly explored and developed. Nevertheless, the concise microhaplotype allelic names have not yet been suggested although there are standardized nomenclatures that were proposed for microhaplotype loci. Here, a proposal was put forward for discussion about microhaplotype alleles being designated with Arabic numerals. For a microhaplotype consisted of single nucleotide polymorphisms (SNPs), the SNPs are locally ordered with their positions in human genome, and have the relevant RefSNP alleles in the dbSNP database be accepted as their possible genotypes. Microhaplotype alleles are allowed to list in every possible combination of the RefSNP alleles before they are arranged in alphabetical order. The ordered alleles are subsequently named with consecutive positive integers starting from 1. Such a nomenclature would be convenient for forensic applications, especially the mixture deconvolution, capable of being enrolled into the software for forensic genetic calculations including PowerStats, Arlequin and STRUCTURE.
Microhaplotypes are combinations of two or more closely linked single nucleotide polymorphisms(SNPs) enclosed within 300 bp DNA fragmental sequence[1, 2, 3]. As an emerging type of genetic marker, microhaplotypes are potential to exert multiple forensic applications, e.g., individual identification[1, 2], family/clan relationship analysis[1, 4], ancestry inference[1, 5, 6, 7, 8, 9] and mixture deconvolution[5, 9, 10].
In 2016, Kidd proposed the nomenclature for microhaplotype loci[11]. Instead of using the complicated rs numbers of SNPs, it suggests that microhaplotypes be named with standardized symbols starting from the letters “mh”followed with chromosome number, lab designation and a lab-specific number. Chen and colleagues introduced a different nomenclature for microhaplotype loci, yet also admitted that they recognized Kidd’ s proposal[12]. A widely acceptant nomenclature and a unique name for each locus will definitely assist microhaplotype research and data handling. However, there have not been suggested concise allelic names for microhaplotypes till present.
This work adopted four DNA samples: two donated ones plus two commercial genomic products: 9947A (Thermo Fisher Scientific, Waltham, MA, USA) and 2800M (Promega, Madison, WI, USA), having been approved by the Ethical Review Board of Institute of Forensic Science, Ministry of Public Security of China. DNA donors have given their written informed consent.
mh21KK-320 amplification was carried out with the forward and reverse primers: 5′ TGACTGGGAGGCTGTGGAGA3′ and 5′ TGCTGGAATTAGAGGCGTGA3′ . Libraries were prepared with 1 ng of inputgenomic DNA to have undergone into the treatmentof TruSeq DNAPCR-Free HT Sample Preparation Kit (Illumina, SanDiego, CA, USA), successively being diluted to 20 pM for a single-run sequencingwith MiSeq ReagentNano Kit (Illumina) on a MiSeqFGx machine (Illumina) so that the reads of 250 bases were thus brought forth. The MHTyper software was selected for microhaplotype allele callingand read counting[13], with the sequencing depth threshold set at 50 reads.
Microhaplotype alleles were here to tentatively designate with Arabic numerals as follows: the forward strand in human genome was to alignmicrohaplotypes; GRCh38, as the most up-to-date sequence assembly until this writing, was therefore to take as the reference sequences; the nomenclature was therewith to: (a) list the genotype-based alleles relating to a microhaplotype locus, (b) establish a unified order of alleles, and (c) nominate alleles with Arabic numerals. To be concrete, SNPs, consisting of a microhaplotype, were to be ordered by their positions in human genome, having the RefSNP alleles in the dbSNP database (https://www.ncbi.nlm.nih.gov/SNP/index.html) accepted as their possible genotypes and all the microhaplotype alleles listed in every possible combination of the RefSNP alleles. The listed microhaplotype alleles were to arrange in alphabetical order, having been named with consecutive positive integers starting from 1. Rare microhaplotype alleles, resulting from SNP genotypes not listed as RefSNP alleles in the database, would still be denoted with the combination of consisting SNP genotypes and placed behind all the numeral-named alleles within the locus.
Microhaplotypes, being combinations of SNPs, are ordinarily straightforward to name their alleles through combining the genotypes of their consisting SNPs, as did in previous publications[1, 12, 14, 15, 16, 17]. When a microhaplotype contains only two SNPs, these genotype-based allelic names are adequate. Whereas, as a microhaplotype consists of more SNPs, the all-elic names will become long, complicated, and even similar to each other. As an example, a simulated DNA mixture was prepared with 9947A plus 2800M and two donated samples R1 and R2 (9947A∶ R1∶ R2∶ 2800M = 1∶ 2∶ 4∶ 8). The five samples (9947A, 2800M, R1, R2 and their mixture made above) were sequenced and genotyped at the locus mh21K-K-320. As illustrated in Fig.1A, the mixture caused trouble to analyze with such genotype-based allelic names even if the sequencing data were fine. It is too complicated to adopt the series of base symbols as a genotype, leaving the allelic information unable to figure out due to failure of credible comparisons into which may take a lot of time and likely lead to mistakes.
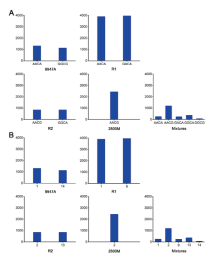 | Fig.1 Histograms of sequencing depths for the locus mh21KK-320, with the alleles being named by bases (A) and digital numbers (B) (Horizontal axis, read counts) |
Multiallelic is an essential advantage of microhaplotypes compared to SNPs. The effective number of alleles (Ae) and informativeness (In) tend to be higher for the microhaplotype loci that cover more SNPs (Fig.2). These loci are more useful in forensic applications, yet the complicated allele names are unacceptable.
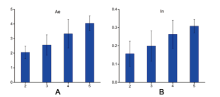 | Fig.2 Histograms of Ae (A) and In (B) for 2~5 microhaplotypes consisting of different numbers of SNPs (Data for 130 microhaplotypes[18] were used for plotting. Error bars: standard deviations) |
Table 1 and 2 listed the respective microhaplotype alleles of locus mh21KK-320 and mh07KK-081 plus their corresponding numeral names, with the latter locus containing an insertion-deletion site. This nomenclature can be utilized to generate unique names for alleles of any microhaplotype locus to be fit after a look-up about the RefSNP alleles of their consisting SNPs in dbSNP database. To make it more convenient, the allelic names for another 130 reported microhaplotypes[18, 19] were presented in supplementary materials (Table S1). Besides, this nomenclature had been already integrated into the software MHTyper of microhaplotype data analysis[13] so that the conversion would be convenient to perform from genotype-based allelic names to the numeral representing ones.
| Table 1 Proposed numeral names of mh21KK-320 alleles |
| Table 2 Proposed numeral names of mh07KK-081 alleles |
According to the nomenclature proposed above, the genotype-based allelic names in Fig.1A were substituted with the numeral allelic ones in Fig.1B. Although it is relatively simple to show the genotype of mh21KK-320 of the four independent samples, as illustrated in Fig. 1A, yet their mixture is time-consuming and confusing to demonstrate with genotype-based allelic names. However, it is facile to recognize the genotypes of each sample as designated with numeral name in Fig. 1B.
Proper and concise names of alleles are essential for forensic genetic markers to exert effective applications. Due to the forensic application of next generation sequencing, STRs have been accepted as sequence-based genetic markers rather than the length-based, having hence brought expectation about the nomenclature for those sequence-based STR alleles and discussed extensively[20]. Microhaplotypes, as one new and promising type of genetic marker, require well-accepted nomenclature for both loci and alleles. In this work, a proposal was put forward on numeral nomenclature for microhaplotype alleles to designate. Its suitability for forensic application stands on the following reasons. Firstly, it avoids the complex combination of SNP genotypes and can be of especial advantages in mixture deconvolution with which convenience would be provided for the relevant forensic application. Secondly, the alleles of each microhaplotype locus are able to be arranged in the order of Arabic numerals. This will offer a unified order of alleles within each locus for data exhibition and communication, as in Fig.1 and Table S1. Thirdly, numeral allelic designation would be easily accepted by forensic scientists because of its similarity to the forensic-available STR alleles. Finally, genetic polymorphisms in different populations are necessary for microhaplotypes to exert efficient forensic applications, among which the current involving bioinformatics software for population genetics, e.g., PowerStats[21], Arlequin[22] and STRUCTURE[23], accepts only numbers as the imported allelic names for statistics[24]. It should be noted that the nomenclature described here is designed for microhaplotypes with already-defined consisting SNPs. When new variants are discovered in an existing microhaplotype sequence, the updated microhaplotype alleles and numeral names should be specified accordingly.
For the supplementary materials of this article, please see: http://www.xsjs-cifs.com/CN/abstract/abstract6830.shtml.
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|


