本文介绍了一种非常灵敏并可应用于法医毒物分析的核酸适配体识别技术。这种技术可与多种分析技术手段联用而检测毒物和药物,技术手段包括阻抗滴定技术、表面增强拉曼技术、荧光检测技术、微流控芯片技术、表面等离子共振技术等。核酸适配体技术的检测优势主要表现为选择性强,并且通过纳米金颗粒修饰,可以极大的增强检测信号,实现对痕量目标分子的精准检测。核酸适配体技术由于其易于合成、造价低廉、操作简便、精准识别等优势必将会受到法庭科学家的重视而广泛应用于未来的法医毒物药物分析工作中。
In this paper, a novel method was introduced and elucidated for the detection of forensic drugs based on nucleic acid aptamers. The method can be combined with diverse analytical technologies, such as the impedimetry, surface-enhanced Raman scattering, fluorescent detection, microfludic sensor and the surface-plasmon resonance. The principle of detection is based on an indirect competitive aptamer-assay which can be greatly enhanced its sensitivity to the concentration-low analytes by resorting to nano-particle coalescence. Aptamers, the molecular recognition agents, are easy to handle for detecting many types of forensically relevant analytes, in particular the drugs. Aptamer-based sensors would fulfill both economical and reliable tasks so that the toxicologists are very hopeful to acquire a time-saving and robust strategic tool for drug analysis.
Drug-related criminal cases, especially those involving in forensic drug abuse, have been increasing in China. These drugs include morphine, heroine, codeine, cocaine and the others of the kind. According to the Ministry of Public Security of the People’ s Republic of China, the drug addicts are more than 14 million by the end of 2015, meaning there was one drug addict every hundred Chinese individuals. To date, drug detection methods have traditionally been based on gas chromatography (GC) and high performance liquid chromatography (HPLC), being time-consuming and expensive to some extent. Several approaches exist on enzymatic immunoassays, yet presenting their disadvantageousness in both the cost and stability of the used enzymes. Aptamers, designed by the SELEX (Systematic Evolution of Ligands by Exponential Enrichment) innovation, were here introduced for detecting forensic drugs. Aptamer technology fulfills the criterion of both economics and stability, thereby being considered a more reliable strategy for drug specific binding.
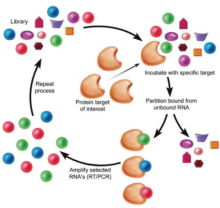
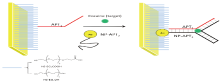
Aptamers are the artificially single-stranded DNA or RNA oligonucleotides, capable of binding to specific molecular targets with high affinity. When integrated to nanomaterials, they may lead to precise detection for forensic drug in common laboratories. In 1990, Tuerk and Gold[1], paralleling at almost the same time, Ellington and Szostsk[2] created a preparation to target the highly specific, high-affinity binding of oligonucleotides. Szostsk named the technology as in-vitro selection while Gold referred to it as SELEX, which was later coined in the literature to represent this technology (Fig.1). This technique is a new combinatorial chemistry, aiming to establish a library of random oligonucleotides and then select a variety of ligands able to bond specifically with oligomeric nucleic acid fragments. One major advantage of the technique is that it has no special requirements for the target molecules. Szostsk first used this technique to select out oligonucleotides and then named it “ aptamer” , derived from the Greek word “ Aptus” , meaning “ to fit” , which afterwards became the generally accepted term. Aptamers are single stranded or double-stranded RNA oligonucleotides and so are the single stranded or double-stranded DNA oligonucleotides. Aptamers are able to form a three dimensional structure, which combines with the bound target molecules at high specificity and high affinity. They also can recognize minute differences between various target molecules, such as distinctions in functional groups (e.g. hydroxyl moiety) or chirality. SELEX has developed aptamers for broad types of target molecules, e.g., small organic molecules[3], sugars[4], amino acids[5], peptides[6], proteins[7], nucleic acids[8] and even cells[9].
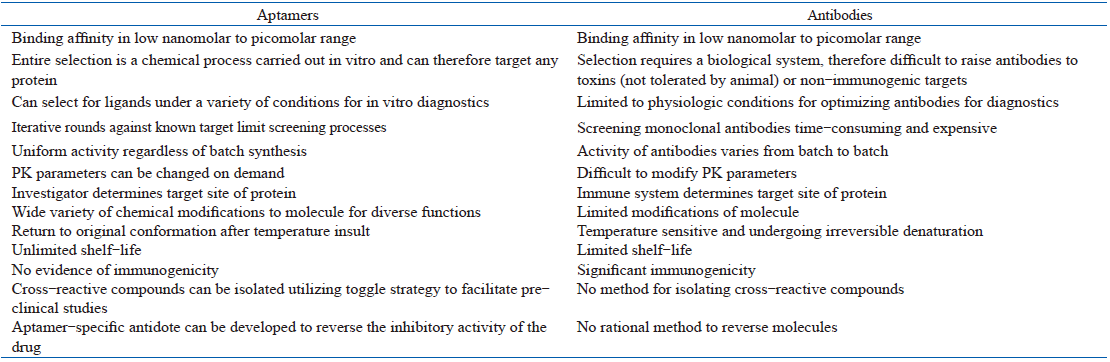
An aptamer’ s recognition mode is similar to that of antibodies but it has the ability to identify similar substances which cannot be discriminated even by monoclonal antibodies. Several properties of aptamers make them attractive recognition-agents that rival, and in some cases surpass, antibodies (Table 1). In addition, aptamers can be synthesized in vitro and have increased stability over antibodies. They can also easily undergo chemical modification and have non-immunogenic advantages, thus enduing them to become one leading and important research tool in many fields.
| Table 1 Properties of aptamers versus antibodies |
Oligonucleotide aptamers have fixed sequences at both the 5’ and 3’ ends of the molecules, yet maintaining the middle section as a random sequence which can bind specifically with the target molecule. Both fixed sequences at the ends are used to expand the oligonucleotide chains, select and prepare the transcription of RNA aptamers. Before a library to be built, the fixed DNA oligonucleotide sequence at each end must first be designed. The length of this fixed sequence is typically among 20-30 nucleotides and contains primer sequences used for PCR amplification and binding sites for restriction endonuclease. The RNA sequences transcribed from the fixed sequences of the DNA oligonucleotide chains must be able to combine with the reverse transcriptase[10]. In a general design, random sequences contain 20-40 nucleotides and can form about 1014-1024 type-different oligonucleotide chains, which can be synthesized by DNA synthesizers (usually by a company). These sequences form chains with a variety of three-dimensional conformations, most of which can be combined with all kinds of target molecules[11]. With the completion of sequence design, the resulting DNA oligonucleotide chains establish the starting DNA oligonucleotide library carrying their random middle sequences (Fig.2).
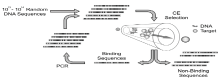
After the preparation of the random oligonucleotide library, the next step is to select and enrich aptamers which can bind to the target molecules with high-specificity and high affinity. Already established methods of selection include nitrocellulose membrane filtration, separation by affinity chromatography, high-speed centrifugation and electrophoresis[12, 13, 14]. Consequently, semi-automatic or automatic and high-throughput SELEX selecting systems have also been founded, decreasing the selection time of aptamers from months to days. The number of repetitions and the simplicity of the process are important indicators in measuring the efficacy of selection and enrichment. Some reported only four rounds of selection and enrichment to obtain aptamers using capillary electrophoresis[15, 16].
Aptamers obtained by the selection and enrichment process need to be analyzed for their base sequence and level II/stereo structure, affinity specificity and lastly, the stability. Sometimes RNA aptamers need to be modified because they are sensitive to RNA enzymes and very easily hydrolyzed. This decreases their shelf-life and can shorten the time that RNA aptamers can be used in the body, thus, limiting their application. Modification methods include[17, 18]: 1) Addition of rare alkali bases or chemically modified alkali bases to the aptamer’ s sequence through the addition of the alkali bases to the reaction system at the beginning of synthesizing the oligonucleotide chains. 2) Modification of RNA aptamers at ribose 2 for fluoride, amino and n-oxygen bases or at the 3’ -ends of RNA aptamers for fatty acids to reduce the effect of the nucleic acid circumscribing enzyme. In the process of preparing ring aptamers, the stability of aptamers can increase 100 times after modification.
Codeine is an alkaloid found in opium. It can either be extracted from opium or chemically synthesized by a morphine manufacturer. Upon arrival into the body, the absorption of approximate 10 % of components is being converted into morphine through the body’ s metabolic process. In List II of the single Convention on Narcotic Drugs of 1961, the United Nations regarded codeine as one of less addictive narcotic drugs, however, it is strictly regulated all over the world.
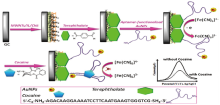
In order to detect codeine, Liangliang Huang et al[19] developed a sensor for opium alkaloid detection employing the codeine-contraposing DNA aptamers generated by the SELEX technique. A novel electrochemical, label-free codeine aptamer-kind biosensor has been proposed by [Fe(CN)6]3-/4- as the electroactive redox probe based on Au-mesoporous silica nanoparticles (Au-MSN) as immobilized substrate. The detection limit of 3 pM was found and the linear range from 10 pM to 100 nM with a correlation coefficient of 0.9979. The result demonstrates that the biosensor is of good stability, regeneration and specificity, capable of being used to detect codeine. Zahra Hashemian et al[20] reported a thin-film microextraction method for the analysis of codeine in urine samples using an aptamer whose immobilization is based on the covalent linkage of an amino-modified anti-codeine aptamer to aldehyde groups of the oxidized cellulose paper. The infrared spectroscopy and elemental analysis were used to examine covalent bonds. The elution conditions (solvent type and volume), extraction time, and extraction temperature were important indicators to measure the extraction efficiency. The results prove that the linear range covers between 10– 300 ng/mL and the detection limit is 3.4 ng/mL in urine.
Methamphetamine (METH) is a highly-addictive synthetic stimulant which causes the body to produce strong pleasant feelings whose effects are sustained for a longtime. For these reasons, it has become a widely-used illicit drug around the world since the year 2000. Fast and cost-effective sensing/detection of METH may help the health authorities to control its abuse, however, such a sensing system is not available yet due to the main lack of suitable METH-sensing probes.
Ebrahimi et al[21] selected an ssDNA aptamer of high binding affinity and specificity to METH through screening a synthetic ssDNA library utilizing SELEX technology (Fig. 3). The specific aptamer was extracted from a large and diverse random ssDNA initial oligonucleotide library with methamphetamine-modified epoxy-activated sepharose 6B. The selected aptamers (aptaMETH) showed high affinity to the target molecule METH with a 100-nM detection limit which can be efficiently distinguish METH from other molecules of similar structures. The results demonstrated the aptaMETH is of specificity and high affinity against methamphetamine, thus resulting in it as a suitable probe for development of METH-sensing aptasensors. Qiunan Shi et al[22] developed a colorimetric method for detection of METH in human urine based on the finding that the presence of METH causes AuNPs solution’ s color changing from red to blue. Usually, aptamers were attached to the surface of AuNP in order to increase their resistance to NaCl-induced aggregation. Urinary METH could be either visually quantified via this effect or by measurement of the absorbance ratios at 660 and 525 nm. The sensor was shown to function within covered concentration ranging from 2 μ M to 10 μ M and its detection limit at 0.82 μ M. The method is believed to represent a widely applicable aptamer-based detection scheme because of its fast and also well-working performance in human urine.
Cocaine is a central nervous system stimulant drug extracted from the coca leaf. From1985, cocaine became the world’ s major drug due to refining technology used to enhance the purity of cocaine. It has several routes of administration (e.g. oral, nasal) and is usually taken at 25-150 mg/dose. Its main effect is a sense of excitement which is experienced within few seconds to a few minutes upon administration.
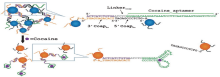
Juewen Liu et al[23] developed a method to detect cocaine based on colorimetric changes (Fig.4). The rate of color change increased with cocaine concentration ascending so that cocaine could be quantified in the range between 50µ m and 500 µ m. The results inspired a general design of colorimetric sensors linked by aptamers based on the disassembly of nanoparticle aggregates. The sensors, demonstrating a fast color change, are easy to use. As no special features required on the aptamers, the design should be applicable to any aptamer of choice. This method is so simple that it can find its applications in many different areas, for example, the electronic industry, environmental monitoring, and medical diagnostics. Yuanfu Zhang et al[24] developed a label-free colorimetric method for DNA hybridization detection based on these unique properties of the (+)AuNPs-SWNTs nanohybrids. The response to target DNA concentration is linear between 0.025 mM and 0.5 mM. The detection limit of cocaine is 2 nM. The method offers the advantages of simplicity, cheapness, rapidity and sensitivity based on the specific recognition of aptamer, therefore very promising for extension to detect non-nucleic acid targets.
Xiaoxia Li et al[25] developed an electrochemical aptasensor incorporating a signal enhancement to detect cocaine. Gold nanoparticles were self-assembled onto the surface of a gold electrode through the use of 1, 6-hexanedithiol whose surface was self-assembled by a bifunctional derivative of the 32-base cocaine-binding aptamer with a redox-active ferrocene moiety and a thiol linker group at the terminus of the strand. The result showed that the linear concentration range of cocaine covers between 1.0 µ M and 15.0 µ M and the detection limit is 0.5 µ M. In addition, this work demonstrated that the sensitivity of the aptasensor modified with gold nanoparticles was 10-fold higher compared with that of the aptasensor without modification of gold nanoparticles and the former provided a promising platform for immobilizing aptamer and enhancing the sensitivity. Qinglin Sheng et al[26] set up an ultrasensitive electrochemical cocaine biosensor based on the three-dimensional (3D) DNA structure conversion of nanostructure from triangular pyramid frustum (TPFDNA) to equilateral triangle (ETDNA). The presence of cocaine triggered the aptamer-composed DNA nanostructure changing from “ close” to “ open” , leading to obvious faradaic impedance changes. The result showed the linear cocaine concentration ranges from 1.0 nM to 2.0 μ M and the correlation coefficient is 0.993. The detection limit is 0.21 nM. The 3D DNA nanostructure of excellent stability and specific rigid structure made the biosensing functions stable, sensitive and regenerative, bringing forth the expectation that its distinctive features would make it potentially advantageous for a diversity of applications to put into practice. Mahmoud Roushani et al[27] designed an aptasensor for the detection of cocaine based on the conformational changes of the aptamer-functionalized gold nanoparticles (AuNPs) onto MWCNTs/IL/Chit nanocomposite as the support platform (Fig.5). The 5’ -amine-3’ -AuNP-terminated aptamer was covalently attached to a MWCNTs/IL/Chit nanocomposite. A decreased current of (K3Fe(CN)6) was monitored by differential pulse voltammetry. The calibration curve of cocaine concentration was linear up to 11 μ M in an optimized condition and the detection limit was 100 pM. This aptasensor can be successfully applied for measuring cocaine in human serum.
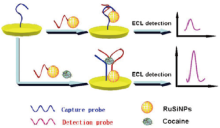
Qihong Cai et al[28] developed an ECL “ sandwich” biosensor to detect cocaine. The sandwich biosensor was fabricated on the basis that a single aptamer could be split into two fragments from which a folded, associated complex could be formed in the presence of the target. In the absence of cocaine, the sensor produced a low ECL signal because of the weak interaction between the two fragments. The association of the two fragments can be induced and stabilized along with the addition of target cocaine into the solution, leading to the immobilization of RuSiNPs onto the electrode surface where to have an enhancing ECL detected. The cocaine concentration covers the range between 1.0× 10-9 and 1.0× 10-11 M and the detection limit is 3.7× 10-12 M. The biosensor was applied to detect trace amounts of cocaine on banknotes with satisfactory results (Fig.6). Tao Li et al[29] proposed a method for the detection of cocaine via nucleic acid aptamers by CE analysis. In order for adaptive binding action to occur, the two aptamers were dissolved in the binding buffer. This result revealed the detection limit was 2 µ M obtained by CE-ECL, comparatively lower than those of previous reports. Yan Li et al[30] also designed an ECL aptamer-based (ECL-AB) biosensor for the determination of a small molecule drugs, using cocaine as a model analyte. The result showed the concentration of cocaine covers the range between 5.0 × 10-9 M and 3.0 × 10-7 M and the detection limit is 1.0 × 10-9 M, demonstrating that this method is a great promising approach for the determination of small molecule drugs.
Dewen Zhang et al[31] designed a label-free method for sensitive and specific detection of cocaine based on the formation of a supramolecular aptamer fragments/substrate complex by electrochemical impedance spectra (EIS). An anti-cocaine aptamer was divided into two fragments, Cx and Cy. Au/Cx5S/MCE, Au/Cy3S/MCE and Au/Cy5S/MCE, were fabricated by immobilizing Cx or Cy on a gold electrode through modifying their 5’ or 3’ end with a thiolated group followed by the treatment with mercaptoethanol (MCE) (Fig.7). In the presence of [Fe(CN)6]3-/4-, the formation of aptamer fragments/cocaine complex was investigated by monitoring electrochemical impedance spectra. The result turned out that Au/Cx5S/MCE had good sensitivity within a detection range from 0.1 µ M to 20 µ M of cocaine. Furthermore, this method improved the sensitivity of the aptasensor significantly. Even without the help of amplification or labeling, cocaine concentrations as low as 100 nM could be easily detected by the developed impedimetric aptasensor. The aptasensor was successfully applied to detect the cocaine in biological fluids. Etery Sharon et al[32] developed methods of electrical impedimetric and field-effect transistor (FET) for analyzing cocaine. The methods were based on the assembly of supramolecular complexes consisting of the aptamer fragments and the respective analyte on electrodes or on the gate of FET devices. The formation of the supramolecular structures enhanced the electronic transduction of the recognition events. While the amplified impedimetric method that employed the nucleic-acid-functionalized AuNPs enhanced the analysis of cocaine with a detection limit of 1 × 10-5 M, the ISFET device elevated the analysis of cocaine with a detection limit of 1 × 10-6 M in the presence of the amplifying AuNPs.
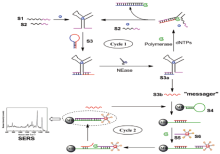
Jiwei Chen et al[33] reported a reagent-less aptameric sensor for the detection of cocaine by surface-enhanced Raman scattering (SERS) spectroscopy with “ signal-on” architecture. This new aptameric sensor was based on the conformational change of the surface-tethered aptamer on a binding target which draws a certain Raman reporter in close to the SERS substrate, thus increasing the Raman scattering signal due to the local enhancement effect of SERS. The sensor can detect cocaine at a concentration of 1 µ m under optimized assay conditions, which compares favorably with analogous aptameric sensors based on electrochemical and fluorescence techniques. The sensor can be readily regenerated by being washed with a buffer. This result showed that the SERS-based transducer might create a new dimension for future development of aptameric sensors for sensitive detection in biochemical and biomedical fields. Ying Li et al[34] proposed a proximity-dependent isothermal cycle amplification (PDICA) strategy which was successfully used for the detection of cocaine coupled with SERS (Fig.8). In order to enhance the SERS signal, Raman dye molecules modified with bio-barcode DNA and AuNPs were applied to prepare the Raman probes. Two label-free proximity probes hybridized with each other and subsequently opened the hairpin connector-probe to perform the PDICA reaction in the presence of target molecules, with inclusion of the amplifications of both strand-displacement and the target recycling. The result demonstrated that the detection limit is 0.1 nM. This assay also showed high selectivity and has been applied to human serum successfully, confirming the reliability and practicality of this protocol.
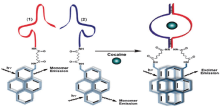
Ronit Freeman et al[35] provided a method for the optical (fluorescence) or electrochemical detection of cocaine via the self-assembly of labeled aptamer sub-units. One of the sub-units is linked to CdSe/ZnS quantum dots (QDs), and the self-assembly of the dye-functionalized second sub-unit with the modified QDs, in the presence of cocaine, stimulates Fö rster resonance energy transfer (FRET). The formation of a supramolecular aptamer-substrate complex allosterically stabilizes the formation of excimer supramolecular structure, and its characteristic emission is observed (Fig.9). In addition, the thiolated aptamer sub-unit is assembled on an Au-electrode. The methylene blue-labeled sub-unit binds to the surface-confined fragment in the presence of cocaine. The amperometric response of the system allows the detection of cocaine in the range of 1× 10-5 M. Cuichen Wu et al [36] reported a method to tailor aptamer sequence into functional subunits for designing target-induced light switching excimer sensors to detect cocaine. The aptamer fragments self-assemble, forming the aptamer-target complex in the presence of target molecules, thus bringing the two pyrene molecules into a close proximity to form an excimer, leading to fluorescent switching from 400 nm to 485 nm. This method has a long fluorescence lifetime, time-resolved measurement, able to directly detect cocaine as low as 5 µ M in urine samples quantitatively without any sample pretreatment. Cuiping Ma et al[37] developed a sensitive fluorescence biosensor for the determination of cocaine based on aptamer and rolling circle amplification. The short DNA strand was displaced from the aptamer, in the presence of cocaine, contributing to cocaine specifically binding with the aptamer. The detection limit was in the range of as low as 0.48 nM. This new method reduced background signal, likely providing a platform for numerous proteins and small molecules to excellently and sensitively detect by DNA amplification.
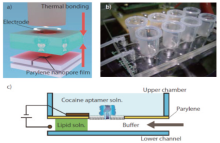
R. Kawano et al[38] described a cocaine sensing using a membrane protein channel integrated in a microfluidic device (Fig.10). This sensor utilizes artificial planar lipid bilayers (BLMs) with α -hemolysin transmembrane protein which allows ssDNA to pass through the application of an electric field. In the unfolded state, ssDNA is attached to a cocaine aptamer spanning across the membrane. The aptamer-cocaine complex no longer fitted through the pore upon binding to cocaine and changing conformation. Thus, the cocaine was discriminated by the protein channel as the structure changing from a cocaine molecule to a cocaine aptamer complex. The detection limit of cocaine was determined to be in the range of 10 µ M in 25 seconds using this system. He Zhang et al[39] reported the development of an aptamer-mediated microfluidic beads-based sensor for cocaine detection based on strand displacement induced by target-aptamer complex. Microbeads functionalized with the aptamers and modified electron of rich proteins were arrayed within a microfluidic channel and were connected with the horseradish peroxidases (HRP) and capturing DNA probe derivative AuNPs by hybridization. The conformational transition of aptamer induced by target-aptamer complex contributes to the displacement of functionalized AuNPs and decreases the fluorescence signal of microbeads. The developed aptamer-based microfluidic bead array sensor was as low as 0.5 pM of cocaine based on the dual signal amplification strategy. This work showed the aptamer-based microfluidic beads array sensor could be used for detection of important molecules in biomedical fields.
Juan Zhang et al[40] designed a bioassay strategy to detect cocaine based on gold nanoparticles (AuNPs) and engineered DNA aptamers with naked eyes. In the presence of the specific target, an aptamer was engineered to be two pieces of random, coil-like single-stranded DNA molecules, which reassemble into the intact aptamer in a tertiary structure. AuNPs can effectively distinguish between these two states via their characteristic surface-plasmon resonance-based color change. Even the concentration of aptamer probes was reduced from 10 to 1 µ M, AuNPs were still well-stabilized and could detect cocaine at 2 µ M. Eyal Golub et al[41] developed a new method that uses metallic or semiconductor nanoparticles (NPs) as labels to detect cocaine by surface plasmon resonance (SPR). The aptasensors are based on the use of two anti-cocaine aptamers’ subunits, where one subunit is assembled on an Au support (acting as an electrode or an SPR-active surface) and the second aptamer subunit is labeled with Pt-NPs, CdS-NPs, or Au-NPs. In different aptasensor configurations, the addition of cocaine results in the formation of supramolecular complexes between the NPs-labeled aptamer subunits and cocaine on the metallic surface, allowing for the quantitative analysis of cocaine. The detection limit was in the range of 10-6 to 10-5 M. The major advantage of this method is free from background interfering signals.
Our research is focused on developing surface chemistry and detection components for rapid, on-site analysis of drugs and other forensically relevant analytes (e.g. TNT) to be commercially or privately used by law enforcement[42, 43]. The current work is to build an aptamer-based biosensor for the detection of TNT (explosive) by a surface-plasmon resonance platform. For this, a novel gold nanoparticle technology is employed to enhance the TNT detection by the aptamer conjugating to the nanoparticles. The principle of detection is based on an indirect competitive aptamer-assay that a greater sensitivity would be further enhanced to the concentration-low analyte by the use of nanoparticle technology. Also, other projects underway include the design of aptamer-based cocaine and morphine sensors resorting to gold nanoparticles for SPR sensing technology to join in (Fig. 11).
In this paper, the novel methods were introduced for the detection of forensic drugs based on nucleic acid aptamers which can be combined with diverse analytes. Technologies of the impedimetry, surface-enhanced Raman scattering, fluorescence-based detection, microfludic sensor and surface-plasmon resonance, can be coalesced into the aptamer approach to play their greater effect. Taking advantage of the excellent selectivity and high affinity of aptamers, the detection will be more accurate and sensitive for the forensic evidence to be decisive. Furthermore, not only are aptamers molecular recognition elements but also economical, easy to handle, capable of designing to detect many types of forensic-purpose analytes.
The authors have declared that no competing interests exist.
作者已声明无竞争性利益关系。
| [1] |
|
| [2] |
|
| [3] |
|
| [4] |
|
| [5] |
|
| [6] |
|
| [7] |
|
| [8] |
|
| [9] |
|
| [10] |
|
| [11] |
|
| [12] |
|
| [13] |
|
| [14] |
|
| [15] |
|
| [16] |
|
| [17] |
|
| [18] |
|
| [19] |
|
| [20] |
|
| [21] |
|
| [22] |
|
| [23] |
|
| [24] |
|
| [25] |
|
| [26] |
|
| [27] |
|
| [28] |
|
| [29] |
|
| [30] |
|
| [31] |
|
| [32] |
|
| [33] |
|
| [34] |
|
| [35] |
|
| [36] |
|
| [37] |
|
| [38] |
|
| [39] |
|
| [40] |
|
| [41] |
|
| [42] |
|
| [43] |
|